Download this page as a Matlab LiveScript
Table of contents
Numerical Integration
We want to integrate some function
We can take any range and map it to the range using iso-parametric mapping:
and its derivative (or map) then we have
Let's say we want to integrate some polynomial of order
Ok, so this can be used to integrate e.g.,
But in general it is awkward to use this rule to integrate e.g.,
or without additional preparation which makes it cumbersome to implement in a computer program and run efficiently.
The same goes for the integration rulse such as
and
Let's consider a polynomial function for
clear
f = @(x) 1+3*x.^2-x.^3;
a = 0; b = 3;
figure
fplot(f,[a,b])
axis([a,b,0,5.1]); box off;
legend('show','Location','Northwest');
We can integrate this analytically
I_e = int(f,sym("x"),a,b)I_e = double(I_e)I_e = 9.7500We can also use the numerical integration using the build in variable precision arithmatic integration
vpaintegral(f,sym("x"),a,b)This uses a numerical method for integration, is powerful and easy to use, drawback are a default high number of function evaluations.
Now, let's go back to basics and take a look at the Riemann sum.
N = 5;
xnod = linspace(a,b,N);
y = double(f(xnod));
figure
fplot(f,[a,b])
axis([a,b,0,5.1]); box off;
legend('show','Location','Northwest');hold on
hb = bar(xnod,y,1,'LineStyle','-',DisplayName="Riemann Sum");
dx = (b-a)/N;
I_R = sum(y*dx);
ht = title(['$I\approx$ ', sprintf('%0.2f', I_R),', $I = $', sprintf(' %0.2f', I_e),', $N=$ ',sprintf(' %d', N)]);
ht.Interpreter = "latex";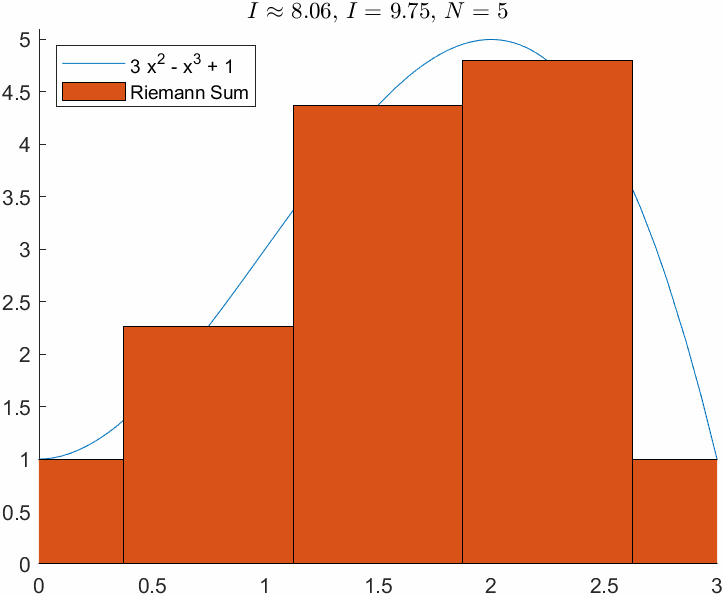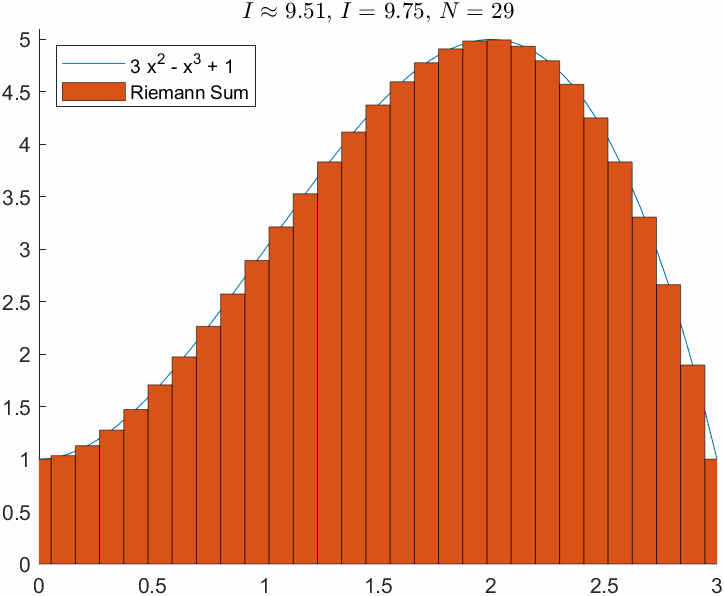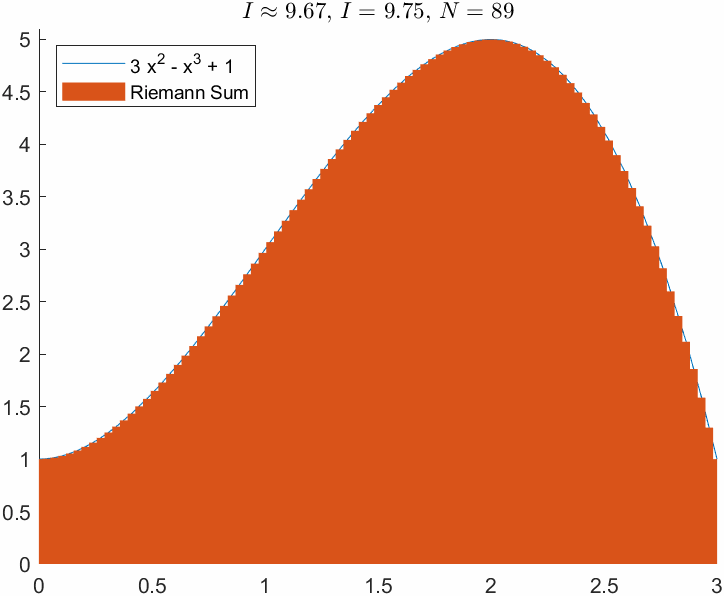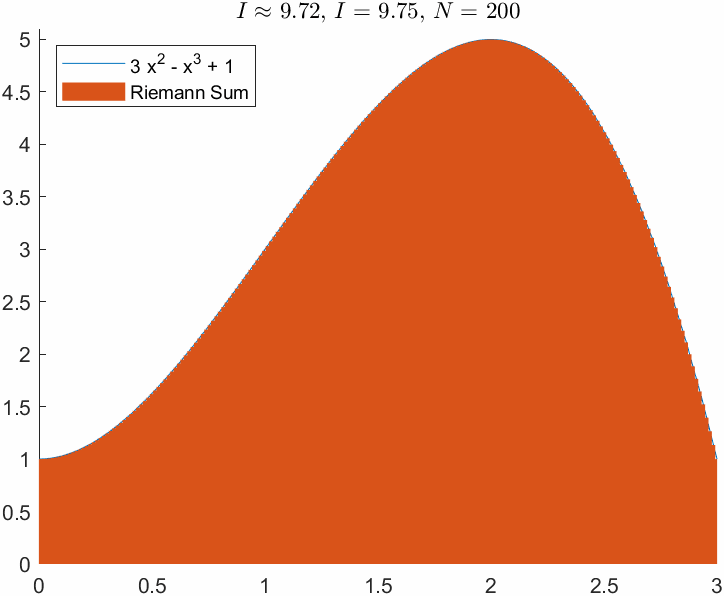
Let's take a look at the error we're making numerically
N = 200;
xnod = linspace(a,b,N);
dx = (b-a)/N;
I_R = sum(f(xnod)*dx);
err_R = abs(I_R-I_e)err_R = 0.0339Note that the absolute integration error is still significantly large after 200 function evaluations!
We can do a better numerical approximation
First we approximate linearly: then we have
This is called the trapezoidal rule. And for a chained approximation with evenly spaced points the approximation becomes:
For example for the function above using we have:
N = 9;
xnod = linspace(a,b,N+1)';
I_R = double(sum(f(xnod)*(b-a)/(N+1)))I_R = 9.0000err_R = abs(I_R-I_e)err_R = 0.7500xnod = linspace(a,b,N+1)';
x1 = xnod(1:end-1);
x2 = xnod(2:end);
I_T = sum(f(x1)+f(x2))*(b-a)/(2*N)I_T = 9.6667I_T2 = trapz(xnod,f(xnod)) % Matlab built in trapezoidal integralI_T2 = 9.6667err_T = double(abs(I_T-I_e))err_T = 0.0833err_R/err_Tans = 9.00009 times better approximation!
figure
axis([a,b,0,5.1]); box off;
legend('show','Location','Northwest');
hold on
nodes = [(1:N)', (2:N+1)'];
xm = mean(xnod(nodes),2);
if N < 15
for iel = 1:N
x1 = xnod(nodes(iel,1));
x2 = xnod(nodes(iel,2));
h = (b-a)/N;
y1 = f(x1);
y2 = f(x2);
area([x1,x2],[y1, y2],'FaceColor','c','EdgeColor','k','HandleVisibility','off');
plot([x1,x1],[0,y1],'k:','HandleVisibility','off')
plot([x2,x2],[0,y2],'k:','HandleVisibility','off')
end
text(xm,xm*0+0.2,cellstr(num2str([1:N]')))
else
area(xnod,f(xnod),'FaceColor','c','HandleVisibility','off');
end
fplot(f,[a,b],'r','LineWidth',2)
plot(xnod,f(xnod),'ok','HandleVisibility','off')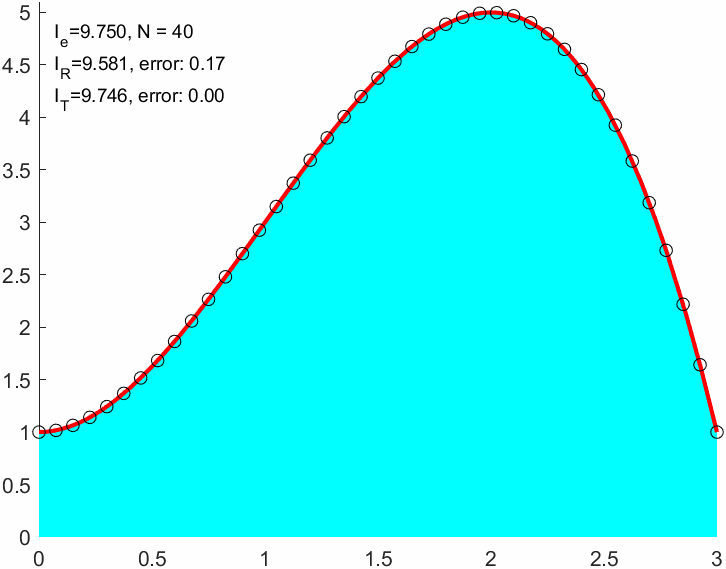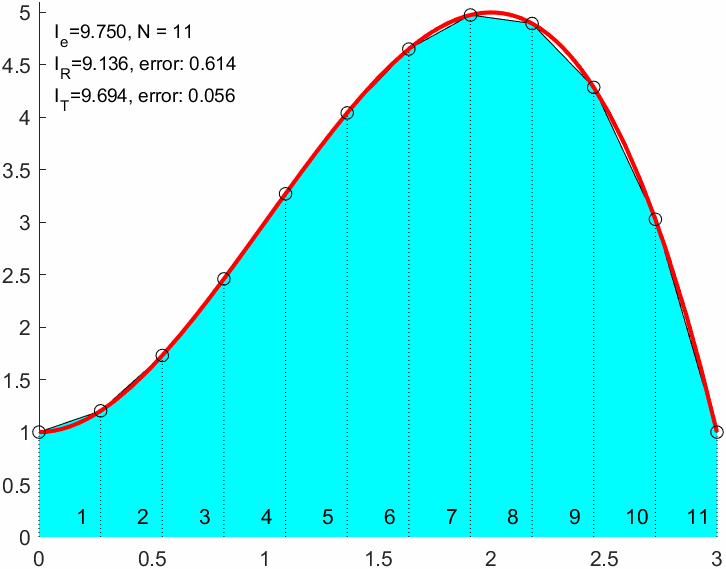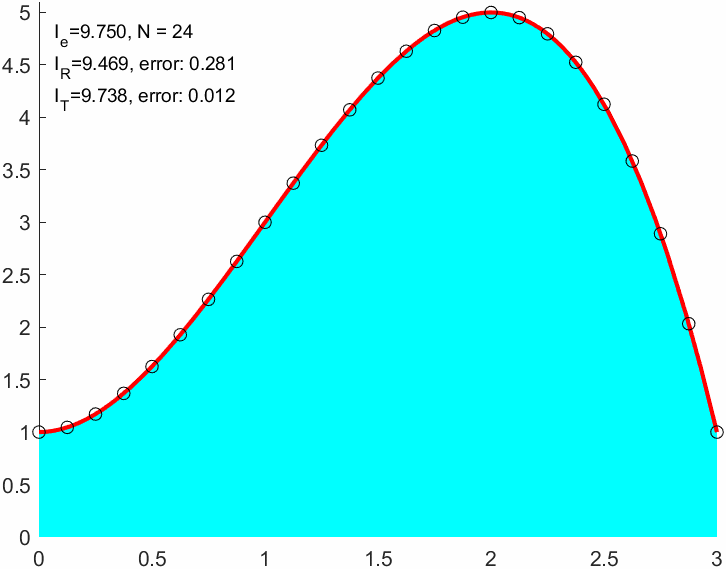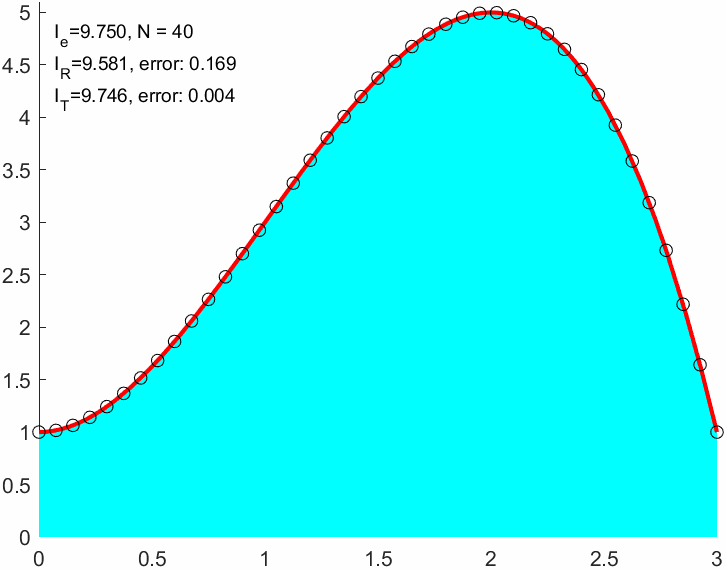
The trapezoidal rule integrates linear polynomials exactly:
f = @(x)1+3*x;
a = 0; b = 3;
I1 = (f(a)+f(b))*(b-a)/2I1 = 16.5000I_exact = vpa(int(sym(f),a,b),5)
But there is a cheaper way...
We can evaluate the midpoint instead:
xm = (a+b)/2;
I1 = (b-a)*f(xm)I1 = 16.5000One function evaluation instead of 2!

This is exact because the error above the graph cancels the error below the graph!
One rule to rule them all.


Gauss quadrature states for
Example:
syms x
p = @(x) x;
I_e = int(sym(p),x,0,1)gp = 1/2; gw = 1;
I_G = p(gp)*gwI_G = 0.5000Ok, not a big deal, after all, even the Riemann sum is exact for linear polynomials.
How about a third order polynomial using only two function evaluations?
where and and
p = @(x)1+3*x.^2-3*x.^3;
gp = [1/2-sym(sqrt(3))/6, 1/2+sym(sqrt(3))/6]gw = sym([1/2, 1/2])I_Gauss = p(gp(1))*gw(1) + p(gp(2))*gw(2)vpa(I_Gauss)simplify(I_Gauss)xnod = linspace(0,1,50);
I_trapz = vpa(trapz(xnod,p(xnod)))I_exact = int(sym(p),x,0,1)figure; hold on
area([0,1/2],[1,1]*p(gp(1)),'FaceColor','c');
area([1/2,1],[1,1]*p(gp(2)),'FaceColor','c');
fplot(p,[0,1], 'r', 'LineWidth',2)
plot(gp,p(gp),'k*')
plot([0.5, 0.5], [0, p(gp(2))], '-k')
The figure visualizes the Gauss integration rule in action. The cyan and white areas between the curves are cancelling out!
Higher order Gauss integration will not make the integral any more accurate, the computed value will be the same. We will just be wasting computational power.
Note that Gauss is not going to exactly integrade other functions in general.
Let's say we need to integrate the function
The resulting integral
is non-elementary.
clear
syms x
f(x) = 1/sqrt(sin(x))
figure
fplot(f,[0,1],'k','LineWidth',2)
axis([0,1,0,10])
xnod = linspace(0.01,1,1000);
ynod = double(f(xnod));
hold on
area(xnod,ynod,'FaceColor','c','EdgeColor','none')
int(f)int(f,x,0,1)We can of course use the built-in numerical integration to solve this
I_e = vpaintegral(f,x,0,1)We can apply Gauss integration using the tabulated values from above:
f = matlabFunction(f);
[gp, gw] = GaussTable(3);
I_G = f(gp)*gwI_G = 1.7857error_Gauss = I_e-I_GWe can see that 3 Gauss points is not enough to sufficiently integrate this function. We need more points!
Let's go down the rabit hole, there exists quadrature tables containing Gauss points for the typical range but there are also several algorithms for computing Gauss points for any Gauss order. Well use the well known Golub-Welsh algorithm taken from L. N. Trefethen, Spectral Methods in Matlab, SIAM, 2000 p.129
f = @(x)1./sqrt(sin(x))
a = 0; b = 1;
N = 17;
[gp, gw] = gauss(N, a, b);
I_G = vpa((b-a)*f(gp)*gw)error_Gauss = abs(I_e-I_G)xnod = linspace(a+0.01,b,N)';
I_T = trapz(xnod,f(xnod))I_T = 1.9312error_Trapz = abs(I_e-I_T)We can plot the error versus the number of evaluations
a = 0; b = 1;
err_G = []; err_Trapz = [];
NP = [2,4,8,16,32,64,128,256];
for N = NP
[gp, gw] = gauss(N, a, b);
I_G = vpa((b-a)*f(gp)*gw);
xnod = linspace(a+0.003,b,N)';
I_T = trapz(xnod,f(xnod));
error_Gauss = abs(I_e-I_G);
error_Trapz = abs(I_e-I_T);
err_G(end+1) = error_Gauss;
err_Trapz(end+1) = error_Trapz;
end
figure;
plot(NP,err_G,'-o','DisplayName','Gauss error'); hold on
plot(NP,err_Trapz,'-x','DisplayName','Trapz error')
gca().set(XScale='log',YScale='log')
legend show
xlabel('N evaluation points')
ylabel('Absolute error')
grid on
xticks(NP)
xticklabels({'2','4','8','16','32','64','128','256'})
We can see that both methods converge to the numerical integral computed by vpaintegral. The trapezoidal integral here is volotile, its lower bound needs to be adjusted to avoid the obious error division by zero. The computation thus depends on the choice of the new lower bound, and for a careful choice of lower bound we can get the trapezoidal method to outperform Gauss for some intermediate number of evaluations. The problem is that we cannot know this value ahead of time and thus the method is not very suitable for this situation.
If we want to apply Gaussian quadrature here, we need to make a variable change to integrate over ranges other that .
We can take any range and map it to the range using iso-parametric mapping:
and its derivative (or map) then we have
and using Gauss quadrature approximation we have
clear
syms x
u(x) = 2*x*sin(2*pi*x)+3
a = -0.3; b = 0.9;
figure
fplot(u,[a,b],'k','LineWidth',2)
% axis([a,b,0,10])
xnod = linspace(a,b,1000);
ynod = double(u(xnod));
hold on
area(xnod,ynod,'FaceColor','c','EdgeColor','none')
I_e = int(u,x,a,b);
I_e = vpa(I_e)And now let's approximate it using Gauss quadrature
f = matlabFunction(u);
N = 8;
[gp, gw] = gauss(N, a, b);
I_G = vpa((b-a)*f(gp)*gw)error_Gauss = I_e-I_Gxnod = linspace(a,b,N)';
I_T = vpa(trapz(xnod,f(xnod)))error_Trapz = I_e-I_TError analysis
error.N = [2,4,8,16,32,64,128,256].';
error.Gauss = zeros(length(error.N),1);
error.Trapz = zeros(length(error.N),1);
for i = 1:length(error.N)
N = error.N(i);
xnod = linspace(a,b,N)';
I_Trapz = trapz(xnod,f(xnod)); % Matlab built in trapezoidal integral
[gp, gw] = gauss(N, a, b);
I_Gauss = (b-a)*f(gp)*gw;
error_Gauss = abs(I_e-I_Gauss);
error_Trapz = abs(I_e-I_Trapz);
error.Gauss(i) = error_Gauss;
error.Trapz(i) = error_Trapz;
end
figure; hold on
plot(error.N, error.Gauss,'-o')
plot(error.N, error.Trapz,'-o')
grid on
set(gca,'xscale','log','yscale','log')
legend('Gauss','Trapz')
xlabel('N evaluation points')
ylabel('Absolute error')
xticks(error.N)
xticklabels({'2','4','8','16','32','64','128','256'})
Here we see that Gauss reaches the same precision as vpaintegral after 16 function evaluations.
Here is a visualization of the Gauss integration method for the function
for
We can integrate the definite integral with limits by the approximation
Now the Gaussian quadrature rule works by evaluating the function at some special points and multiplying by some special weights , the linear combination will result in an approximate integral that for a certain polynomial is exact as integration points integrate polynomials of degree exactly!
When is not a polynomial function, we still can approximate the integral by in a sense choosing the closest matching polynomial of the type and the corresponding Gauss quadrature rule to integrate this polynomial exactly.
These special points and weights can be found by many ways where the Golub-Welsch algorithm is implemented in the gauss.m function, but we shall here take a look at another which is arguably simpler to explain.
For a certain approximation, e.g., the linear
we have
with
This yields a system of equations
This also works for higher order approximations, e.g., cubic. Note we need polynomial functions which contain even terms since the points and weights are associated with the unknown coefficients
with
from which we form
Now we could solve this symbolically, below, but we shall also see how to solve this system of non-linear equations using numerically using Newton's method.
syms w1 w2 x1 x2 x
ekv = [w1 + w2 == int(1,x,0,1)
w1*x1 + w2*x2 == int(x,x,0,1)
w1*x1^2 + w2*x2^2 == int(x^2,x,0,1)
w1*x1^3 + w2*x2^3 == int(x^3,x,0,1)]
[w1 w2 x1 x2] = solve(ekv, [w1 w2 x1 x2])We get two solutions:
The Newton method states
where
Now we rewrite this such that we can turn it into a linear system
or
with , and
We will also need initial values for , we will choose randomly such that they all are in the interval .
syms w1 w2 x1 x2 x
f1 = w1 + w2 - int(1,x,0,1);
f2 = w1*x1 + w2*x2 - int(x,x,0,1);
f3 = w1*x1^2 + w2*x2^2 - int(x^2,x,0,1);
f3 = w1*x1^2 + w2*x2^2 - int(x^2,x,0,1);
J = [diff(f1,w1), diff(f1, w2), diff(f1, x1), diff(f1, x2)
diff(f2,w1), diff(f2, w2), diff(f2, x1), diff(f2, x2)
diff(f3,w1), diff(f3, w2), diff(f3, x1), diff(f3, x2)
diff(f4,w1), diff(f4, w2), diff(f4, x1), diff(f4, x2)];
f = [f1
f2
f3
f4];
% Going numeric here
J = matlabFunction(J);
f = matlabFunction(f);
w1 = rand; w2 = rand; x1 = rand; x2 = rand;
xold = [w1
w2
x1
x2];
for i = 1:1000
xold = [w1
w2
x1
x2];
Jnew = J(w1,w2,x1,x2);
fnew = f(w1,w2,x1,x2);
xd = Jnew\-fnew;
xnew = xold + xd;
w1 = xnew(1); w2 = xnew(2); x1 = xnew(3); x2 = xnew(4);
eps = norm(xold - xnew)
if eps < 1e-12 %stop at an arbitrary precision
break;
end
endDepending on the initial values, the algorithm will take a different amount of iterations to converge below the set tolerance.
eps = 0.5225
eps = 0.0709
eps = 0.0195
eps = 6.9134e-04
eps = 2.6636e-07
eps = 1.1066e-13The converged numerical values:
w1 = 0.5000
w2 = 0.5000
x1 = 0.2113
x2 = 0.7887function [gp, gw] = GaussTable(n)
switch n
case 1
gp = 0.5;
gw = 1;
case 2
gp = [0.5-sqrt(3)/6
0.5+sqrt(3)/6]';
gw = [0.5
0.5];
case 3
gp = [0.5-sqrt(3)*sqrt(5)/10
0.5
0.5+sqrt(3)*sqrt(5)/10]';
gw = [5/18
4/9
5/18];
end
endfunction [x, w, A] = gauss(n, a, b)
%------------------------------------------------------------------------------
% gauss.m
%------------------------------------------------------------------------------
%
% Purpose:
%
% Generates abscissas and weights on I = [ a, b ] (for Gaussian quadrature).
%
% Implementation of the Golub-Welsch algorithm
% https://en.wikipedia.org/wiki/Gaussian_quadrature#The_Golub-Welsch_algorithm
%
% Syntax:
%
% [x, w, A] = gauss(n, a, b);
%
%
% Input:
%
% n integer Number of quadrature points.
% a real Left endpoint of interval.
% b real Right endpoint of interval.
%
%
% Output:
%
% x real Quadrature points.
% w real Weigths.
% A real Area of domain.
%------------------------------------------------------------------------------
% 3-term recurrence coefficients:
n = 1:(n - 1);
beta = 1 ./ sqrt(4 - 1 ./ (n .* n));
% Jacobi matrix:
J = diag(beta, 1) + diag(beta, -1);
% Eigenvalue decomposition:
%
% e-values are used for abscissas, whereas e-vectors determine weights.
%
[V, D] = eig(J);
x = diag(D);
% Size of domain:
A = b - a;
% Change of interval:
%
% Initally we have I0 = [ -1, 1 ]; now one gets I = [ a, b ] instead.
%
% The abscissas are Legendre points.
%
if ~(a == -1 && b == 1)
x = 0.5 * A * x + 0.5 * (b + a);
end
% Weigths:
w = V(1, :) .* V(1, :);
w = w';
x=x';
end